fit
Description
The fit function fits a configured one-class support vector
machine (SVM) model for incremental anomaly detection (incrementalOneClassSVM object) to streaming data.
To fit a one-class SVM model to an entire batch of data at once, see ocsvm.
Mdl = fit(Mdl,Tbl)Mdl, which represents the input incremental learning model Mdl trained using the predictor data in Tbl.
Specifically, the fit function fits the model to the incoming
data and stores the updated score threshold and configurations in the output model
Mdl.
[
additionally returns the numeric array Mdl,tf,scores] = fit(___)scores containing anomaly scores
with N elements for N observations. The values in
this array are in the range (–Inf,Inf). A negative score value with large
magnitude indicates a normal observation, and a large positive value indicates an
anomaly.
Examples
Create a default one-class support vector machine (SVM) model for incremental anomaly detection.
Mdl = incrementalOneClassSVM; Mdl.ScoreWarmupPeriod
ans = 0
Mdl.ContaminationFraction
ans = 0
Mdl is an incrementalOneClassSVM model object. All its properties are read-only. By default, the software sets the score warm-up period to 0 and the anomaly contamination fraction to 0.
Mdl must be fit to data before you can use it to perform any other operations.
Load Data
Load the 1994 census data stored in census1994.mat. The data set consists of demographic data from the US Census Bureau.
load census1994.matincrementalOneClassSVM does not support categorical predictors and does not use observations with missing values. Remove missing values in the data to reduce memory consumption and speed up training. Remove the categorical predictors.
adultdata = rmmissing(adultdata); adultdata = removevars(adultdata,["workClass","education","marital_status", ... "occupation","relationship","race","sex","native_country","salary"]);
Fit Incremental Model
Fit the incremental model Mdl to the data in the adultdata table by using the fit function. Because ScoreWarmupPeriod = 0, fit returns scores and detects anomalies immediately after fitting the model for the first time. To simulate a data stream, fit the model in chunks of 100 observations at a time. At each iteration:
Process 100 observations.
Overwrite the previous incremental model with a new one fitted to the incoming observations.
Store
medianscore, the median score value of the data chunk, to see how it evolves during incremental learning.Store
allscores, the score values for the fitted observations.Store
threshold, the score threshold value for anomalies, to see how it evolves during incremental learning.Store
numAnom, the number of detected anomalies in the data chunk.
n = numel(adultdata(:,1)); numObsPerChunk = 100; nchunk = floor(n/numObsPerChunk); medianscore = zeros(nchunk,1); threshold = zeros(nchunk,1); numAnom = zeros(nchunk,1); allscores = []; % Incremental fitting rng(0,"twister"); % For reproducibility for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; Mdl = fit(Mdl,adultdata(idx,:)); [isanom,scores] = isanomaly(Mdl,adultdata(idx,:)); medianscore(j) = median(scores); allscores = [allscores scores']; numAnom(j) = sum(isanom); threshold(j) = Mdl.ScoreThreshold; end
Mdl is an incrementalOneClassSVM model object trained on all the data in the stream. The fit function fits the model to the data chunk, and the isanomaly function returns the observation scores and the indices of observations in the data chunk with scores above the score threshold value.
Analyze Incremental Model During Training
Plot the anomaly score for every observation.
plot(allscores,".-") xlabel("Observation") ylabel("Score") xlim([0 n])

At each iteration, the software calculates a score value for each observation in the data chunk. A negative score value with large magnitude indicates a normal observation, and a large positive value indicates an anomaly.
To see how the score threshold and median score per data chunk evolve during training, plot them on separate tiles.
figure tiledlayout(2,1); nexttile plot(medianscore,".-") ylabel("Median Score") xlabel("Iteration") xlim([0 nchunk]) nexttile plot(threshold,".-") ylabel("Score Threshold") xlabel("Iteration") xlim([0 nchunk])

finalScoreThreshold=Mdl.ScoreThreshold
finalScoreThreshold = 0.1799
The median score is negative for the first several iterations, then rapidly approaches zero. The anomaly score threshold immediately rises from its (default) starting value of 0 to 1.3, and then gradually approaches 0.18. Because ContaminationFraction = 0, incrementalOneClassSVM treats all training observations as normal observations, and at each iteration sets the score threshold to the maximum score value in the data chunk.
totalAnomalies = sum(numAnom)
totalAnomalies = 0
No anomalies are detected at any iteration, because ContaminationFraction = 0.
Train a one-class SVM model on a simulated noisy periodic shingled time series containing no anomalies by using ocsvm. Convert the trained model to an incremental learner object, and incrementally fit the time series and detect anomalies.
Create Simulated Data Stream
Create a simulated data stream of observations representing a noisy sinusoid signal.
rng(0,"twister"); % For reproducibility period = 100; n = 5001+period; sigma = 0.04; a = linspace(1,n,n)'; b = sin(2*pi*(a-1)/period)+sigma*randn(n,1);
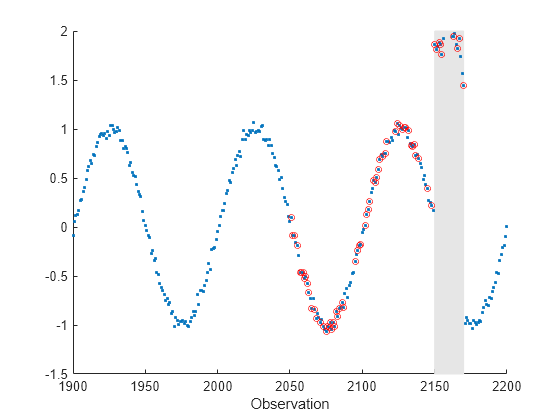
Introduce an anomalous region into the data stream. Plot the data stream portion which contains the anomalous region, and circle the anomalous data points.
c = 2*(sin(2*pi*(a-35)/period)+sigma*randn(n,1));
b(2150:2170) = c(2150:2170); scatter(a,b,".") xlim([1900,2200]) xlabel("Observation") hold on scatter(a(2150:2170),b(2150:2170),"r") hold off

Convert the single-featured data set b into a multi-featured data set by shingling [1] with a shingle size equal to the period of the signal. The th shingled observation is a vector of features with values , , ..., , where is the shingle size.
X = []; shingleSize = period; for i = 1:n-shingleSize X = [X;b(i:i+shingleSize-1)']; end
Train Model and Perform Incremental Anomaly Detection
Fit a one-class SVM model to the first 1000 shingled observations, specifying a contamination fraction of zero. Convert it to an incrementalOneClassSVM model object.
Mdl = ocsvm(X(1:1000,:),ContaminationFraction=0); IncrementalMdl = incrementalLearner(Mdl);
To simulate a data stream, process the full shingled data set in chunks of 100 observations at a time. At each iteration:
Process 100 observations.
Calculate scores and detect anomalies using the
isanomalyfunction.Store
anomIdx, the indices of shingled observations marked as anomalies.If the chunk contains fewer than three anomalies, fit and update the previous incremental model.
n = numel(X(:,1)); numObsPerChunk = 100; nchunk = floor(n/numObsPerChunk); anomIdx = []; allscores = []; % Incremental fitting rng(0,"twister"); % For reproducibility for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; [isanom,scores] = isanomaly(IncrementalMdl,X(idx,:)); allscores = [allscores;scores]; anomIdx = [anomIdx;find(isanom)+ibegin-1]; if (sum(isanom) < 3) IncrementalMdl = fit(IncrementalMdl,X(idx,:)); end end
Analyze Incremental Model During Training
At each iteration, the software calculates a score value for each observation in the data chunk. A negative score value with large magnitude indicates a normal observation, and a large positive value indicates an anomaly. Plot the anomaly score for the observations in the vicinity of the anomaly. Circle the scores of shingles that the software returns as anomalous.
figure scatter(a(1:5000),allscores,".") hold on scatter(a(anomIdx),allscores(anomIdx),20,"or") xlim([1900,2200]) xlabel("Shingle") ylabel("Score") hold off

Because the introduced anomalous region begins at observation 2150, and the shingle size is 100, shingle 2051 is the first one to show a high anomaly score. Some shingles between 2050 and 2170 have scores lying just below the anomaly score threshold due to the noise in the sinusoidal signal. The shingle size affects the performance of the model by defining how many subsequent consecutive data points in the original time series the software uses to calculate the anomaly score for each shingle.
Plot the unshingled data and highlight the introduced anomalous region. Circle the observation number of the first element in each shingle that the software returned as anomalous.
figure xlim([1900,2200]) ylim([-1.5 2]) rectangle(Position=[2150 -1.5 20 3.5],FaceColor=[0.9 0.9 0.9], ... EdgeColor=[0.9 0.9 0.9]) hold on scatter(a,b,".") scatter(a(anomIdx),b(anomIdx),20,"or") xlabel("Observation") hold off
Train a one-class SVM model and perform anomaly detection on a data set with categorical predictors.
Load Data
Load the 1994 census data stored in census1994.mat. The data set consists of demographic data from the US Census Bureau.
load census1994.matThe fit function of incrementalOneClassSVM does not use observations with missing values. Remove missing values in the data to reduce memory consumption and speed up training.
adultdata = rmmissing(adultdata); adulttest = rmmissing(adulttest);
The census data set contains nine categorical variables. Because the fit function of incrementalOneClassSVM does not support categorical variables, you need to convert them to dummy variables. Remove all of the noncategorical variables, and remove the categorical variables that have more than 10 unique categories. Convert the remaining categorical variables to dummy variables using onehotencode.
adultdata = removevars(adultdata,["age","fnlwgt","capital_gain", ... "capital_loss","hours_per_week","occupation","education", ... "education_num","native_country"]); adulttest = removevars(adulttest,["age","fnlwgt","capital_gain", ... "capital_loss","hours_per_week","occupation","education", ... "education_num","native_country"]); Xtrain = table(); Xstream = table(); for i=1:width(adultdata) Xtrain = [Xtrain onehotencode(adultdata(:,i))]; Xstream = [Xstream onehotencode(adulttest(:,i))]; end
Train One-Class SVM Model
Fit a one-class SVM model to the training data. Specify a random stream for reproducibility, and an anomaly contamination fraction of 0.001. Set KernelScale to "auto" so that the software selects an appropriate kernel scale parameter using a heuristic procedure.
rng(0,"twister"); % For reproducibility TTMdl = ocsvm(Xtrain,ContaminationFraction=0.001, ... KernelScale="auto",RandomStream=RandStream("mlfg6331_64"))
TTMdl =
OneClassSVM
CategoricalPredictors: []
ContaminationFraction: 1.0000e-03
ScoreThreshold: -0.6840
PredictorNames: {1×30 cell}
KernelScale: 2.4495
Lambda: 0.0727
Properties, Methods
TTMdl is a OneClassSVM model object representing a traditionally trained one-class SVM model.
Convert Trained Model
Convert the traditionally trained one-class SVM model to a one-class SVM model for incremental learning.
IncrementalMdl = incrementalLearner(TTMdl);
IncrementalMdl is an incrementalOneClassSVM model object that is ready for incremental learning and anomaly detection.
Fit Incremental Model and Detect Anomalies
Perform incremental learning on the Xstream data by using the fit function. To simulate a data stream, fit the model in chunks of 100 observations at a time. At each iteration:
Process 100 observations.
Overwrite the previous incremental model with a new one fitted to the incoming observations.
Store
medianscore, the median score value of the data chunk, to see how it evolves during incremental learning.Store
threshold, the score threshold value for anomalies, to see how it evolves during incremental learning.Store
numAnom, the number of detected anomalies in the chunk, to see how it evolves during incremental learning.
n = numel(Xstream(:,1)); numObsPerChunk = 100; nchunk = floor(n/numObsPerChunk); medianscore = zeros(nchunk,1); numAnom = zeros(nchunk,1); threshold = zeros(nchunk,1); % Incremental fitting for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; [IncrementalMdl,tf,scores] = fit(IncrementalMdl,Xstream(idx,:)); medianscore(j) = median(scores); numAnom(j) = sum(tf); threshold(j) = IncrementalMdl.ScoreThreshold; end
Analyze Incremental Model During Training
To see how the median score, score threshold, and number of detected anomalies per chunk evolve during training, plot them on separate tiles.
tiledlayout(3,1); nexttile plot(medianscore) ylabel("Median Score") xlabel("Iteration") xlim([0 nchunk]) nexttile plot(threshold) ylabel("Score Threshold") xlabel("Iteration") xlim([0 nchunk]) nexttile plot(numAnom,"+") ylabel("Anomalies") xlabel("Iteration") xlim([0 nchunk]) ylim([0 max(numAnom)+0.2])
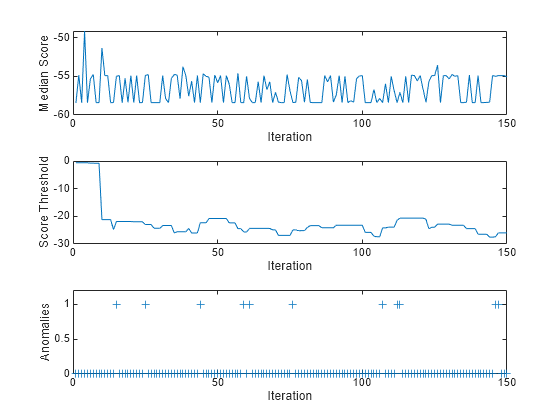
totalanomalies=sum(numAnom)
totalanomalies = 11
anomfrac= totalanomalies/n
anomfrac = 7.3041e-04
fit updates the model and returns the observation scores and the indices of observations with scores above the score threshold value as anomalies. A negative score value with large magnitude indicates a normal observation, and a large positive value indicates an anomaly. The median score fluctuates between approximately 58 and 55. After the 10th iteration, the score threshold fluctuates between 28 and 21. The software detects 11 anomalies in the Xstream data, yielding a total contamination fraction of approximately 0.0007.
Input Arguments
Incremental anomaly detection model to fit to streaming data, specified as an
incrementalOneClassSVM model object. You can create Mdl
by calling incrementalOneClassSVM directly, or by converting a
traditionally trained OneClassSVM
model using the incrementalLearner function.
Predictor data, specified as a table. Each row of Tbl
corresponds to one observation, and each column corresponds to one predictor variable.
Multicolumn variables and cell arrays other than cell arrays of character vectors are
not allowed.
If you train Mdl using a table, then you must provide predictor
data by using Tbl, not X. All predictor
variables in Tbl must have the same variable names and data types
as those in the training data. However, the column order in Tbl
does not need to correspond to the column order of the training data.
Note
If an observation contains at least one missing value (
NaN,''(empty character vector),""(empty string),<missing>, or<undefined>) ,fitignores the observation. Consequently,fituses fewer than n observations to create an updated model, where n is the number of observations inTbl.Incremental learning functions support only numeric input predictor data. You must prepare an encoded version of categorical data to use incremental learning functions. Use
dummyvarto convert each categorical variable to a dummy variable. For more details, see Dummy Variables.
Data Types: table
Predictor data, specified as a numeric matrix. Each row of X
corresponds to one observation, and each column corresponds to one predictor
variable.
If you train Mdl using a matrix, then you must provide
predictor data by using X, not Tbl. The
variables that make up the columns of X must have the same order as
the columns in the training data.
Note
If an observation contains at least one missing (
NaN) value,fitignores the observation. Consequently,fituses fewer than n observations to create an updated model, where n is the number of observations inX.Incremental learning functions support only numeric input predictor data. You must prepare an encoded version of categorical data to use incremental learning functions. Use
dummyvarto convert each categorical variable to a numeric matrix of dummy variables. Then, concatenate all dummy variable matrices and any other numeric predictors, in the same way that the training function encodes categorical data. For more details, see Dummy Variables.
Data Types: single | double
Output Arguments
Updated one-class SVM model for incremental anomaly detection, returned as an
incrementalOneClassSVM model object.
Anomaly indicators, returned as a logical column vector. An element of
tf is true when the observation in the
corresponding row of Tbl or X is an anomaly,
and false otherwise. tf has the same length as
Tbl or X.
fit updates Mdl and then detects
observations with scores above the threshold (the
ScoreThreshold value) as anomalies.
Note
If the model is not warm (
IsWarm=false), thenfitreturns alltfasfalse.fitassigns the anomaly indicator offalse(logical 0) to observations with at least one missing value.
Data Types: logical
Anomaly scores, returned as a numeric column vector whose values are in the range
(–Inf,Inf). scores has the same length as
Tbl or X, and each element of
scores contains an anomaly score for the observation in the
corresponding row of Tbl or X. fit
calculates scores after updating Mdl. A negative score value with
large magnitude indicates a normal observation, and a large positive value indicates an
anomaly.
Note
If the model is not warm (
IsWarm=false), thenfitreturns allscoresasNaN.fitassigns the anomaly score ofNaNto observations with at least one missing value.
Data Types: single | double
References
[1] Guha, Sudipto, N. Mishra, G. Roy, and O. Schrijvers. "Robust Random Cut Forest Based Anomaly Detection on Streams," Proceedings of The 33rd International Conference on Machine Learning 48 (June 2016): 2712–21.
Version History
Introduced in R2023b
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Seleccione un país/idioma
Seleccione un país/idioma para obtener contenido traducido, si está disponible, y ver eventos y ofertas de productos y servicios locales. Según su ubicación geográfica, recomendamos que seleccione: .
También puede seleccionar uno de estos países/idiomas:
Cómo obtener el mejor rendimiento
Seleccione China (en idioma chino o inglés) para obtener el mejor rendimiento. Los sitios web de otros países no están optimizados para ser accedidos desde su ubicación geográfica.
América
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)