rnaplot
Draw secondary structure of RNA sequence
Syntax
Description
rnaplot( draws the RNA secondary
structure specified by RNA2ndStruct)RNA2ndStruct, the secondary structure of an RNA
sequence represented by a string scalar or character vector specifying bracket notation or a
connectivity matrix.
ha = rnaplot(RNA2ndStruct)ha, a handle to the figure axis.
[ also returns ha,H]
= rnaplot(RNA2ndStruct)H, a
structure of handles, which you can use to graph elements in a MATLAB® figure window.
___ = rnaplot(
specifies options using one or more name-value arguments in addition to the input arguments
in previous syntaxes. For example, to color by residue type, set
RNA2ndStruct,Name=Value)ColorBy to "Residue".
Examples
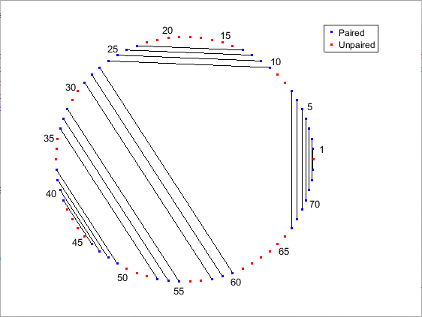
Determine the minimum free-energy secondary structure of an RNA sequence and plot it in circle format.
seq = "GCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAAAGGUUAGGGGUUCGACUCCUCUCGGGCGCG";
RNA2ndStructure = rnafold(seq);
rnaplot(RNA2ndStructure)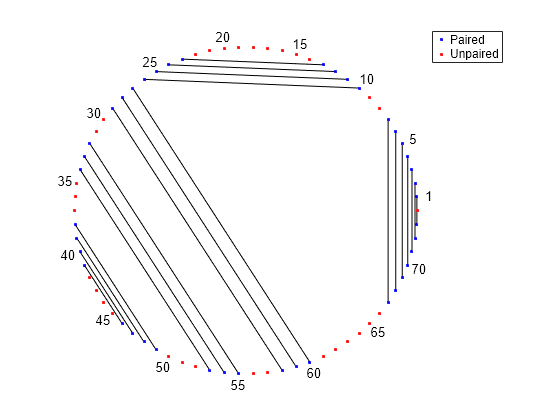
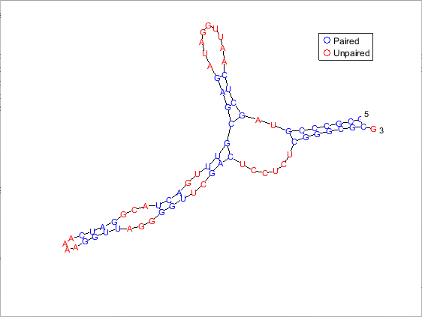
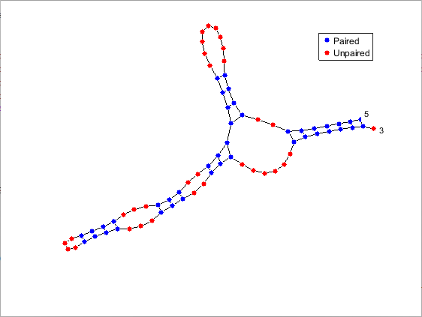
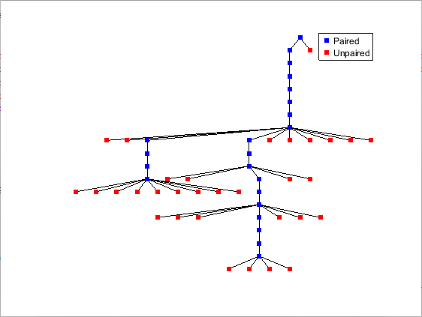
Plot the RNA sequence secondary structure in graph format and color the plot by pair type.
seq = "GCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAAAGGUUAGGGGUUCGACUCCUCUCGGGCGCG"; RNA2ndStructure = rnafold(seq); rnaplot(RNA2ndStructure,Sequence=seq,Format="Graph",ColorBy="Pair")
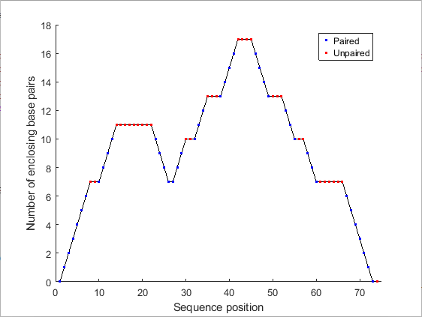
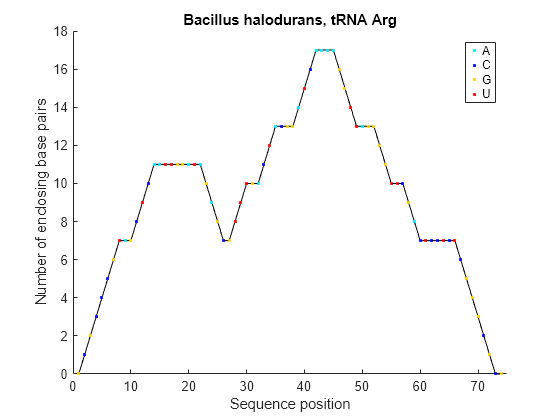
Plot the RNA sequence secondary structure in mountain format and color the plot by residue type. Use the handle ha to add a title to the plot.
seq = "GCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAAAGGUUAGGGGUUCGACUCCUCUCGGGCGCG"; RNA2ndStructure = rnafold(seq); ha = rnaplot(RNA2ndStructure,Sequence=seq,Format="Mountain",ColorBy="Residue"); title(ha,"Bacillus halodurans, tRNA Arg")
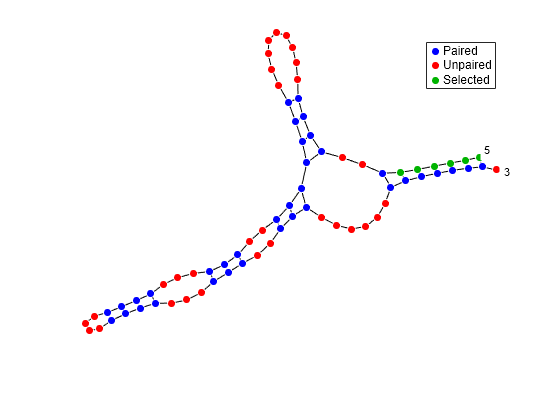
Mutate the first six positions in an RNA sequence and observe the effect the mutation has on the secondary structure by highlighting the first six residues.
seq = "GCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAAAGGUUAGGGGUUCGACUCCUCUCGGGCGCG"; seqMut = replaceBetween(seq,1,6,"AAAAAA"); RNA2ndStructure = rnafold(seq); RNA2ndStructureMut = rnafold(seqMut); rnaplot(RNA2ndStructure,Sequence=seq,Format="DotDiagram",Selection=1:6)
rnaplot(RNA2ndStructureMut,Sequence=seqMut,Format="DotDiagram",Selection=1:6)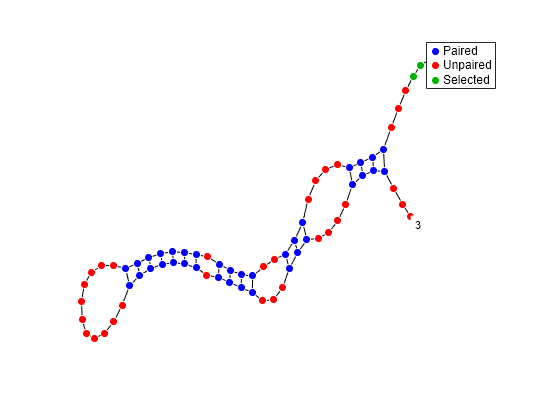
Input Arguments
Secondary structure of RNA sequence, specified as a string scalar, character vector,
or binary matrix. Use bracket notation to specify RNA2ndStruct as a
string scalar or character vector, and use a binary matrix to specify
RNA2ndStruct as a connectivity matrix.
Tip
You can use the rnafold function to create
RNA2ndStruct.
Data Types: string | char | logical | double | single | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: rnaplot(RNA2ndStructure,Selection="Unpaired")
RNA secondary structure sequence to plot, specified as a string scalar, character
vector, or structure containing a Sequence field. The information
in Sequuence is used in the data tip displayed by clicking a base
in the plot of the RNA secondary structure RNA2ndStruct.
Example: "GCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAAAGGUUAGGGGUUCGACUCCUCUCGGGCGCG"
Data Types: string | char | struct
Plot format, specified as "Circle",
"Diagram", "Dotdiagram",
"Graph", "Mountain", or
"Tree". The choices are described in this table.
| Format | Description |
|---|---|
"Circle" | Each base is represented by a dot on the circumference of a circle. Lines connect bases that pair with each other.
|
"Diagram" | Each base is represented and identified by a letter. The backbone and hydrogen bonds between base pairs are represented by lines.
Note If you specify |
"Dotdiagram" | Each base is represented and identified by a dot. The backbone and hydrogen bonds between base pairs are represented by lines.
|
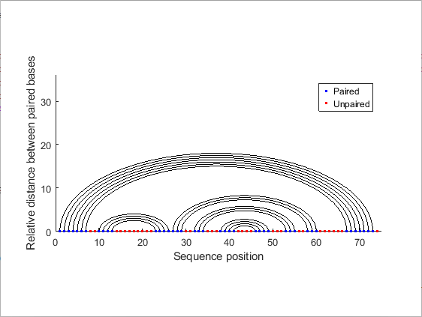
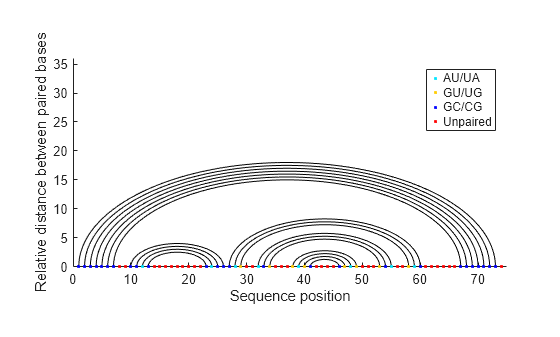
"Graph" | Bases are displayed in their sequence position along the abscissa (horizontal axis) of a graph. Semi-elliptical curves connect bases that pair with each other. The height of the lines is proportional to the distance between paired bases.
|
"Mountain" | Each base is represented by a dot in a two-dimensional plot, where the base position is in the abscissa (horizontal axis) and the number of base pairs enclosing a given base is in the ordinate (vertical axis).
|
"Tree" | Each base is represented by a node in a tree graph. Leaf nodes indicate unpaired bases, while each internal node indicates a base pair. The tree root is a fictitious node, not associated with any base in the secondary structure.
Note If you specify |
Data Types: string | char
Residues to highlight in plot, specified as a numeric vector, a string scalar, or
a character vector. If you specify Selection as a numeric vector,
then the rnaplot function highlights the residues whose indices
appear as entries in Selection. If you specify
Selection as a string scalar or character vector, then the
rnaplot function highlights the subset of residues described by
Selection. In this case, Selection must
have one of these values:
"Paired""Unpaired""AU"(or"UA")"GC"(or"CG")"GU"(or"UG")
Note
If you specify Selection as "AU",
"GC", or "GU" (or "UA",
"CG", or "UG"), then you must also specify a
value for Sequence to provide the RNA sequence.
Data Types: double | single | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64 | string | char
Color scheme for plot, specified as "State",
"Residue", or "Pair". The choices are
described in this list.
"State"— Color by pair state: paired bases and unpaired bases."Residue"— Color by residue type: A, C, G, and U."Pair"— Color by pair type: AU/UA, GC/CG, and GU/UG.
Note
If you specify ColorBy as "Residue" or
"Pair", then you must also specify a value for
Sequence to provide the RNA sequence.
Data Types: string | char
Output Arguments
Handle to axes, returned as an Axes object.
Handles from plot, returned as a structure. Depending on the values you choose for
Selection and ColorBy,
H can contain these fields:
Paired— Handles to all paired residuesUnpaired— Handles to all unpaired residuesA— Handles to all A residuesC— Handles to all C residuesG— Handles to all G residuesU— Handles to all U residuesAU— Handles to all AU/UA pairsGC— Handles to all GC/CG pairsGU— Handles to all GU/UG pairsSelected— Handles to all selected residues
Tip
Use the handles returned in H to change properties of the
graph elements, such as color, marker size, and marker type.
Version History
Introduced in R2007b
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Seleccione un país/idioma
Seleccione un país/idioma para obtener contenido traducido, si está disponible, y ver eventos y ofertas de productos y servicios locales. Según su ubicación geográfica, recomendamos que seleccione: .
También puede seleccionar uno de estos países/idiomas:
Cómo obtener el mejor rendimiento
Seleccione China (en idioma chino o inglés) para obtener el mejor rendimiento. Los sitios web de otros países no están optimizados para ser accedidos desde su ubicación geográfica.
América
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)