seqlogo
Display sequence logo for nucleotide or amino acid sequences
Syntax
Description
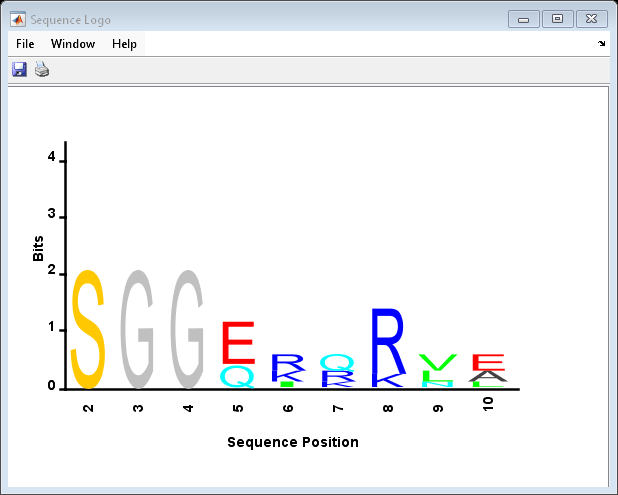
seqlogo( displays a sequence logo for
Seqs)Seqs, a set of aligned sequences. The logo graphically displays the
sequence conservation at a particular position in the alignment of sequences, measured in
bits. The maximum sequence conservation per site is log2(4) bits for
nucleotide sequences and log2(20) bits for amino acid sequences. If the
sequence conservation value is zero or negative, no logo is displayed in that
position.
seqlogo( displays a sequence logo for
Profile)Profile, a sequence profile distribution matrix with the frequency of
nucleotides or amino acids for every column in the multiple alignment, such as returned by
the seqprofile function.
seqlogo(
uses additional options specified by one or more Seqs,Name,Value)Name,Value pair
arguments.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
References
[1] Schneider, T.D., and Stephens, R.M. (1990). Sequence Logos: A new way to display consensus sequences. Nucleic Acids Research 18, 6097–6100.