kfoldPredict
Classify observations in cross-validated kernel classification model
Description
label = kfoldPredict(CVMdl)ClassificationPartitionedKernel) CVMdl. For every fold,
kfoldPredict predicts class labels for validation-fold observations
using a model trained on training-fold observations.
[
also returns classification scores
for both classes.label,score] = kfoldPredict(CVMdl)
Examples
Classify observations using a cross-validated, binary kernel classifier, and display the confusion matrix for the resulting classification.
Load the ionosphere data set. This data set has 34 predictors and 351 binary responses for radar returns, which are labeled either bad ('b') or good ('g').
load ionosphereCross-validate a binary kernel classification model using the data.
rng(1); % For reproducibility CVMdl = fitckernel(X,Y,'Crossval','on')
CVMdl =
ClassificationPartitionedKernel
CrossValidatedModel: 'Kernel'
ResponseName: 'Y'
NumObservations: 351
KFold: 10
Partition: [1×1 cvpartition]
ClassNames: {'b' 'g'}
ScoreTransform: 'none'
Properties, Methods
CVMdl is a ClassificationPartitionedKernel model. By default, the software implements 10-fold cross-validation. To specify a different number of folds, use the 'KFold' name-value pair argument instead of 'Crossval'.
Classify the observations that fitckernel does not use in training the folds.
label = kfoldPredict(CVMdl);
Construct a confusion matrix to compare the true classes of the observations to their predicted labels.
C = confusionchart(Y,label);
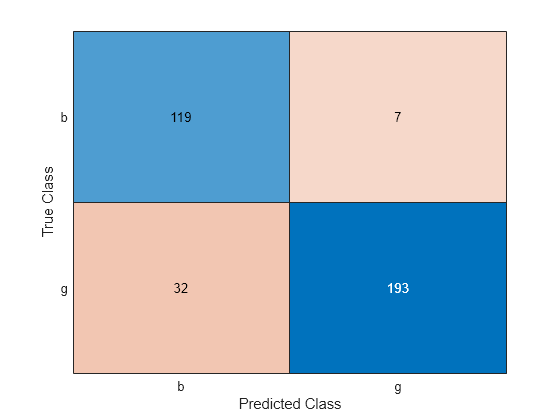
The CVMdl model misclassifies 32 good ('g') radar returns as being bad ('b') and misclassifies 7 bad radar returns as being good.
Estimate posterior class probabilities using a cross-validated, binary kernel classifier, and determine the quality of the model by plotting a receiver operating characteristic (ROC) curve. Cross-validated kernel classification models return posterior probabilities for logistic regression learners only.
Load the ionosphere data set. This data set has 34 predictors and 351 binary responses for radar returns, which are labeled either bad ('b') or good ('g').
load ionosphereCross-validate a binary kernel classification model using the data. Specify the class order, and fit logistic regression learners.
rng(1); % For reproducibility CVMdl = fitckernel(X,Y,'Crossval','on', ... 'ClassNames',{'b','g'},'Learner','logistic')
CVMdl =
ClassificationPartitionedKernel
CrossValidatedModel: 'Kernel'
ResponseName: 'Y'
NumObservations: 351
KFold: 10
Partition: [1×1 cvpartition]
ClassNames: {'b' 'g'}
ScoreTransform: 'none'
Properties, Methods
CVMdl is a ClassificationPartitionedKernel model. By default, the software implements 10-fold cross-validation. To specify a different number of folds, use the 'KFold' name-value pair argument instead of 'Crossval'.
Predict the posterior class probabilities for the observations that fitckernel does not use in training the folds.
[~,posterior] = kfoldPredict(CVMdl);
The output posterior is a matrix with two columns and n rows, where n is the number of observations. Column i contains posterior probabilities of CVMdl.ClassNames(i) given a particular observation.
Compute the performance metrics (true positive rates and false positive rates) for a ROC curve and find the area under the ROC curve (AUC) value by creating a rocmetrics object.
rocObj = rocmetrics(Y,posterior,CVMdl.ClassNames);
Plot the ROC curve for the second class by using the plot function of rocmetrics.
plot(rocObj,ClassNames=CVMdl.ClassNames(2))
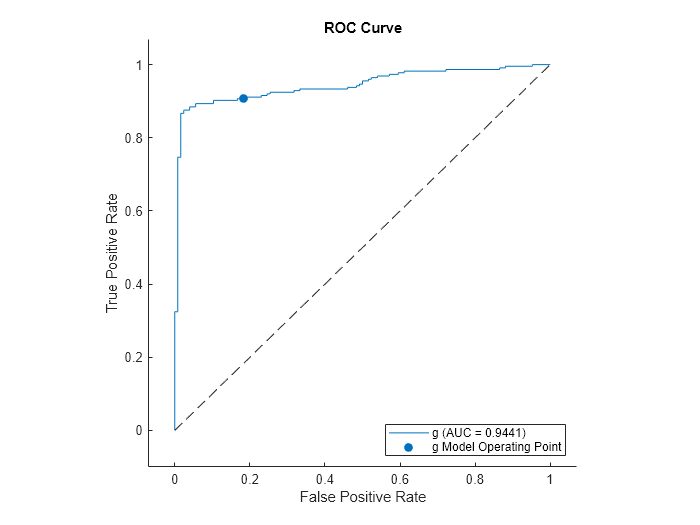
The AUC is close to 1, which indicates that the model predicts labels well.
Input Arguments
Cross-validated, binary kernel classification model, specified as a ClassificationPartitionedKernel model object. You can create a
ClassificationPartitionedKernel model by using fitckernel
and specifying any one of the cross-validation name-value pair arguments.
To obtain estimates, kfoldPredict applies the same data used to
cross-validate the kernel classification model (X and
Y).
Output Arguments
Predicted class labels, returned as a categorical or character array, logical or numeric matrix, or cell array of character vectors.
label has n rows, where n
is the number of observations in X, and has the same data type as the
observed class labels (Y) used to train CVMdl.
(The software treats string arrays as cell arrays of character
vectors.)
kfoldPredict classifies observations into the class yielding
the highest score.
Classification
scores, returned as an n-by-2 numeric array, where
n is the number of observations in X.
score(i,j) is the score for classifying observation
i into class j. The order of the classes is
stored in CVMdl.ClassNames.
If CVMdl.Trained{1}.Learner is 'logistic',
then classification scores are posterior probabilities.
More About
For kernel classification models, the raw classification score for classifying the observation x, a row vector, into the positive class is defined by
is a transformation of an observation for feature expansion.
β is the estimated column vector of coefficients.
b is the estimated scalar bias.
The raw classification score for classifying x into the negative class is −f(x). The software classifies observations into the class that yields a positive score.
If the kernel classification model consists of logistic regression learners, then the
software applies the 'logit' score transformation to the raw
classification scores (see ScoreTransform).
Extended Capabilities
This function fully supports GPU arrays. For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2018bkfoldPredict fully supports GPU arrays.
Starting in R2023b, the following classification model object functions use observations with missing predictor values as part of resubstitution ("resub") and cross-validation ("kfold") computations for classification edges, losses, margins, and predictions.
In previous releases, the software omitted observations with missing predictor values from the resubstitution and cross-validation computations.
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Seleccione un país/idioma
Seleccione un país/idioma para obtener contenido traducido, si está disponible, y ver eventos y ofertas de productos y servicios locales. Según su ubicación geográfica, recomendamos que seleccione: .
También puede seleccionar uno de estos países/idiomas:
Cómo obtener el mejor rendimiento
Seleccione China (en idioma chino o inglés) para obtener el mejor rendimiento. Los sitios web de otros países no están optimizados para ser accedidos desde su ubicación geográfica.
América
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)